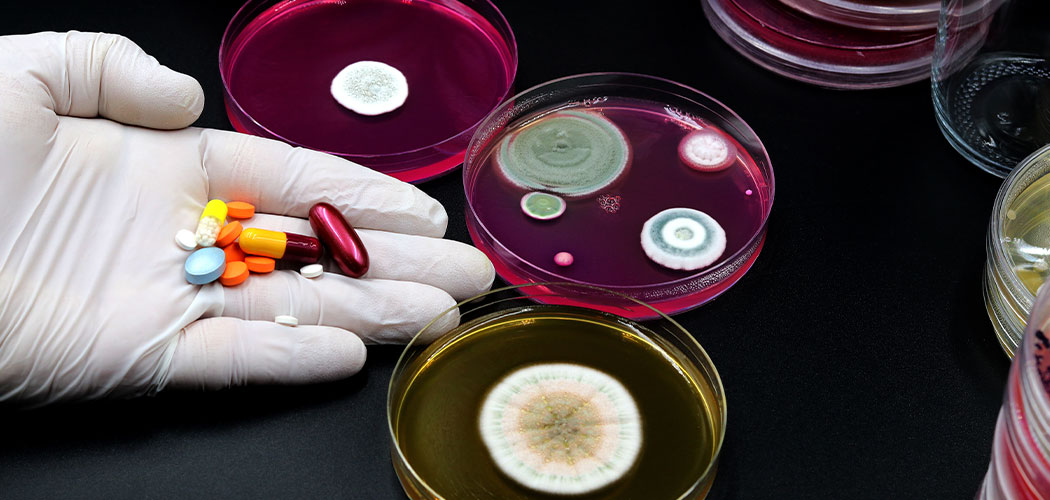
Researchers have used advanced data science to describe and predict when resistance to antibiotics will emerge during treatment for a bacterial infection, based purely on the knowledge of pre-existing bacterial resistance mutations.
The development of medicines to treat infections cannot keep up with the rise in antimicrobial resistance, one of the top global public health threats, researchers say. So, there is a pressing need for new ways to predict the emergence of antibiotic resistance during treatment to ensure the dose is right and the drug is working as effectively as possible.
The researchers, led by Monash University, focused on the antibiotic ‘meropenem’ which is administered intravenously to treat a range of serious bacterial infections such as meningitis, intra-abdominal infection, pneumonia and sepsis.
In the study, published in Clinical Microbiology and Infection, the team developed the first ever quantitative systems pharmacology (QSP) model to describe and predict meropenem resistance across seven strains of the bacterium Pseudomonas aeruginosa, a pathogen associated with antibiotic resistance and high mortality in immunocompromised patients.
QSP uses computational models to describe interactions between a drug and disease. In this case, the QSP model describes and predicts the full time-course of bacterial growth, bacterial killing and antibiotic resistance across each of the seven Pseudomonas aeruginosa strains that had various pre-existing bacterial characteristics, including resistance mutations.
Co-lead author, Associate Professor Cornelia Landersdorfer from the Monash Institute of Pharmaceutical Sciences (MIPS), said the QSP model represents a substantial advancment compared with the way scientists currently attempt to measure the varying activity of antibiotics over time following administration.
“The relationship between antibiotic, bacterial characteristics and resistance emergence during treatment is complicated, which means getting dosing regimens right can be tricky,” Associate Professor Landersdorfer said.
Co-lead author Associate Professor Antonio Oliver from the Instituto de Investigación Sanitaria Illes Balears (IdISBa) and Hospital Son Espases, Palma de Mallorca, Spain said: “To our knowledge, no previous studies have employed an integrated experimental and modelling approach to investigate meropenem regimens in the context of different resistance mechanisms using a panel of bacterial resistant mutants.”
Pseudomonas aeruginosa possesses an exceptional ability for resistance emergence during antibiotic treatment and is among the leading pathogens causing deaths associated with antimicrobial resistance worldwide.
“The antibiotic meropenem is commonly used against Pseudomonas aeruginosa. Traditionally, pharmacokinetic/pharmacodynamic indices have been used in an attempt to link antibiotic concentrations after initiating treatment with bacterial response, and inform dosing. However, this method does not capture the full time-courses of exposure to the antibiotic and the subsequent bacterial response, and does not account for important pre-existing bacterial characteristics, including mutations, that can influence resistance emergence,” said Associate Professor Landersdorfer.
“In contrast, QSP models describe and predict the full time-courses of bacterial growth and resistance emergence.”
Researchers believe the study, funded by an NHMRC Ideas grant and Centre of Research Excellence, provides a potential future approach towards enabling optimised antibiotic therapy that is tailored to the characteristics of the bacterial strain causing an infection.
Read the full study here